Nathan Weeks
2014-04-21 15:51:05 UTC
To augment Scott's response: from a quick reading of the the
Bio::Graphics::Glyph::gene code, it looks like if one specifies a
value for "sub_part", only subfeatures of that type will be rendered
(e.g., if one specifies "sub_part = exon", then UTRs won't be
rendered). Here's the relevant code (I think) in the
Bio::Graphics::Glyph::gene module:
======================================
my @subparts;
if ($class->option('sub_part')) {
@subparts = $feature->get_SeqFeatures($class->option('sub_part'));
}
elsif ($feature->primary_tag =~ /^mRNA/i) {
@subparts = $feature->get_SeqFeatures(qw(CDS five_prime_UTR
three_prime_UTR UTR));
}
else {
@subparts = $feature->get_SeqFeatures('exon');
}
======================================
It would be nice if sub_part accepted a list of types.
--
Nathan Weeks
IT Specialist
USDA-ARS Corn Insects and Crop Genetics Research Unit
Crop Genome Informatics Laboratory
Iowa State University
http://weeks.public.iastate.edu/
On Thu, Apr 17, 2014 at 4:48 PM,
Bio::Graphics::Glyph::gene code, it looks like if one specifies a
value for "sub_part", only subfeatures of that type will be rendered
(e.g., if one specifies "sub_part = exon", then UTRs won't be
rendered). Here's the relevant code (I think) in the
Bio::Graphics::Glyph::gene module:
======================================
my @subparts;
if ($class->option('sub_part')) {
@subparts = $feature->get_SeqFeatures($class->option('sub_part'));
}
elsif ($feature->primary_tag =~ /^mRNA/i) {
@subparts = $feature->get_SeqFeatures(qw(CDS five_prime_UTR
three_prime_UTR UTR));
}
else {
@subparts = $feature->get_SeqFeatures('exon');
}
======================================
It would be nice if sub_part accepted a list of types.
--
Nathan Weeks
IT Specialist
USDA-ARS Corn Insects and Crop Genetics Research Unit
Crop Genome Informatics Laboratory
Iowa State University
http://weeks.public.iastate.edu/
On Thu, Apr 17, 2014 at 4:48 PM,
------------------------------
Message: 7
Date: Thu, 17 Apr 2014 10:47:22 -0700 (PDT)
Subject: [Gmod-gbrowse] Configuring Genes to show exons and UTRs
Content-Type: text/plain; charset=us-ascii
Hello,
I'm having an issue trying to render exons and UTRs together. Here is a
/Chr10 Ensembl_AGPv3 gene 1720611 1722779 . + .
Name=GRMZM2G129907;biotype=protein_coding;description=Uncharacterized%20protein%20%20[Source:UniProtKB/TrEMBL%3BAcc:B4FHQ6];logic_name=genebuilder;ID=GRMZM2G129907
Chr10 Ensembl_AGPv3 mRNA 1720611 1722779 . + .
Name=GRMZM2G129907_T02;Parent=GRMZM2G129907;biotype=protein_coding;description=cdna|est|omrna;logic_name=genebuilder;ID=GRMZM2G129907_T02
Chr10 Ensembl_AGPv3 mRNA 1720611 1722779 . + .
Name=GRMZM2G129907_T01;Parent=GRMZM2G129907;biotype=protein_coding;description=est|omrna|protein;logic_name=genebuilder;ID=GRMZM2G129907_T01
Chr10 Ensembl_AGPv3 mRNA 1721887 1722779 . + .
Name=GRMZM2G129907_T03;Parent=GRMZM2G129907;biotype=protein_coding;description=est|omrna|protein;logic_name=genebuilder;ID=GRMZM2G129907_T03
Chr10 Ensembl_AGPv3 exon 1720611 1720931 . + .
Name=GRMZM2G129907_E02;Parent=GRMZM2G129907_T01;ensembl_end_phase=2;ensembl_phase=-1;rank=1;ID=GRMZM2G129907_E02
Chr10 Ensembl_AGPv3 five_prime_UTR 1720611 1720713 . + .
Parent=GRMZM2G129907_T01;
Chr10 Ensembl_AGPv3 CDS 1720714 1720931 . + 0
Name=GRMZM2G129907_P01;Parent=GRMZM2G129907_T01;rank=1;ID=GRMZM2G129907_P01
Chr10 Ensembl_AGPv3 CDS 1721395 1721501 . + 0
Name=GRMZM2G129907_P01;Parent=GRMZM2G129907_T01;rank=2;ID=GRMZM2G129907_P01
Chr10 Ensembl_AGPv3 exon 1721395 1721501 . + .
Name=GRMZM2G129907_E04;Parent=GRMZM2G129907_T01;ensembl_end_phase=1;ensembl_phase=2;rank=2;ID=GRMZM2G129907_E04
Chr10 Ensembl_AGPv3 CDS 1722296 1722459 . + 0
Name=GRMZM2G129907_P01;Parent=GRMZM2G129907_T01;rank=3;ID=GRMZM2G129907_P01
Chr10 Ensembl_AGPv3 exon 1722296 1722779 . + .
Name=GRMZM2G129907_E03;Parent=GRMZM2G129907_T01;ensembl_end_phase=-1;ensembl_phase=1;rank=3;ID=GRMZM2G129907_E03
Chr10 Ensembl_AGPv3 three_prime_UTR 1722460 1722779 . + .
Parent=GRMZM2G129907_T01;
Chr10 Ensembl_AGPv3 exon 1720611 1721501 . + .
Name=GRMZM2G129907_E06;Parent=GRMZM2G129907_T02;ensembl_end_phase=-1;ensembl_phase=-1;rank=1;ID=GRMZM2G129907_E06
Chr10 Ensembl_AGPv3 five_prime_UTR 1720611 1720713 . + .
Parent=GRMZM2G129907_T02;
Chr10 Ensembl_AGPv3 CDS 1720714 1720935 . + 0
Name=GRMZM2G129907_P02;Parent=GRMZM2G129907_T02;rank=1;ID=GRMZM2G129907_P02
Chr10 Ensembl_AGPv3 three_prime_UTR 1720936 1722779 . + .
Parent=GRMZM2G129907_T02;
Chr10 Ensembl_AGPv3 exon 1721994 1722055 . + .
Name=GRMZM2G129907_E05;Parent=GRMZM2G129907_T02;ensembl_end_phase=-1;ensembl_phase=-1;rank=2;ID=GRMZM2G129907_E05
Chr10 Ensembl_AGPv3 exon 1722296 1722779 . + .
Name=GRMZM2G129907_E01;Parent=GRMZM2G129907_T02;ensembl_end_phase=-1;ensembl_phase=-1;rank=3;ID=GRMZM2G129907_E01
Chr10 Ensembl_AGPv3 exon 1721887 1722055 . + .
Name=GRMZM2G129907_E07;Parent=GRMZM2G129907_T03;ensembl_end_phase=-1;ensembl_phase=-1;rank=1;ID=GRMZM2G129907_E07
Chr10 Ensembl_AGPv3 five_prime_UTR 1721887 1722363 . + .
Parent=GRMZM2G129907_T03;
Chr10 Ensembl_AGPv3 exon 1722296 1722779 . + .
Name=GRMZM2G129907_E01;Parent=GRMZM2G129907_T03;ensembl_end_phase=-1;ensembl_phase=-1;rank=2;ID=GRMZM2G129907_E01
Chr10 Ensembl_AGPv3 CDS 1722364 1722459 . + 0
Name=GRMZM2G129907_P03;Parent=GRMZM2G129907_T03;rank=1;ID=GRMZM2G129907_P03
Chr10 Ensembl_AGPv3 three_prime_UTR 1722460 1722779 . + .
Parent=GRMZM2G129907_T03;/
<Loading Image...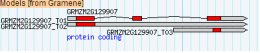 >
>
This is close to what I want, but the inner exons aren't showing. When I use
"sub_part=exon" in this track's stanza, the exons show but the gray UTRs
<Loading Image...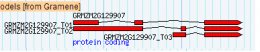 >
>
I want to render a display that looks exactly like the image above, but with
the grayed UTR regions showing in the exons. Any help on accomplishing this
would be greatly appreciated.
Thanks,
John
--
View this message in context: http://generic-model-organism-system-database.450254.n5.nabble.com/Configuring-Genes-to-show-exons-and-UTRs-tp5712467.html
Sent from the gmod-gbrowse mailing list archive at Nabble.com.
Message: 7
Date: Thu, 17 Apr 2014 10:47:22 -0700 (PDT)
Subject: [Gmod-gbrowse] Configuring Genes to show exons and UTRs
Content-Type: text/plain; charset=us-ascii
Hello,
I'm having an issue trying to render exons and UTRs together. Here is a
/Chr10 Ensembl_AGPv3 gene 1720611 1722779 . + .
Name=GRMZM2G129907;biotype=protein_coding;description=Uncharacterized%20protein%20%20[Source:UniProtKB/TrEMBL%3BAcc:B4FHQ6];logic_name=genebuilder;ID=GRMZM2G129907
Chr10 Ensembl_AGPv3 mRNA 1720611 1722779 . + .
Name=GRMZM2G129907_T02;Parent=GRMZM2G129907;biotype=protein_coding;description=cdna|est|omrna;logic_name=genebuilder;ID=GRMZM2G129907_T02
Chr10 Ensembl_AGPv3 mRNA 1720611 1722779 . + .
Name=GRMZM2G129907_T01;Parent=GRMZM2G129907;biotype=protein_coding;description=est|omrna|protein;logic_name=genebuilder;ID=GRMZM2G129907_T01
Chr10 Ensembl_AGPv3 mRNA 1721887 1722779 . + .
Name=GRMZM2G129907_T03;Parent=GRMZM2G129907;biotype=protein_coding;description=est|omrna|protein;logic_name=genebuilder;ID=GRMZM2G129907_T03
Chr10 Ensembl_AGPv3 exon 1720611 1720931 . + .
Name=GRMZM2G129907_E02;Parent=GRMZM2G129907_T01;ensembl_end_phase=2;ensembl_phase=-1;rank=1;ID=GRMZM2G129907_E02
Chr10 Ensembl_AGPv3 five_prime_UTR 1720611 1720713 . + .
Parent=GRMZM2G129907_T01;
Chr10 Ensembl_AGPv3 CDS 1720714 1720931 . + 0
Name=GRMZM2G129907_P01;Parent=GRMZM2G129907_T01;rank=1;ID=GRMZM2G129907_P01
Chr10 Ensembl_AGPv3 CDS 1721395 1721501 . + 0
Name=GRMZM2G129907_P01;Parent=GRMZM2G129907_T01;rank=2;ID=GRMZM2G129907_P01
Chr10 Ensembl_AGPv3 exon 1721395 1721501 . + .
Name=GRMZM2G129907_E04;Parent=GRMZM2G129907_T01;ensembl_end_phase=1;ensembl_phase=2;rank=2;ID=GRMZM2G129907_E04
Chr10 Ensembl_AGPv3 CDS 1722296 1722459 . + 0
Name=GRMZM2G129907_P01;Parent=GRMZM2G129907_T01;rank=3;ID=GRMZM2G129907_P01
Chr10 Ensembl_AGPv3 exon 1722296 1722779 . + .
Name=GRMZM2G129907_E03;Parent=GRMZM2G129907_T01;ensembl_end_phase=-1;ensembl_phase=1;rank=3;ID=GRMZM2G129907_E03
Chr10 Ensembl_AGPv3 three_prime_UTR 1722460 1722779 . + .
Parent=GRMZM2G129907_T01;
Chr10 Ensembl_AGPv3 exon 1720611 1721501 . + .
Name=GRMZM2G129907_E06;Parent=GRMZM2G129907_T02;ensembl_end_phase=-1;ensembl_phase=-1;rank=1;ID=GRMZM2G129907_E06
Chr10 Ensembl_AGPv3 five_prime_UTR 1720611 1720713 . + .
Parent=GRMZM2G129907_T02;
Chr10 Ensembl_AGPv3 CDS 1720714 1720935 . + 0
Name=GRMZM2G129907_P02;Parent=GRMZM2G129907_T02;rank=1;ID=GRMZM2G129907_P02
Chr10 Ensembl_AGPv3 three_prime_UTR 1720936 1722779 . + .
Parent=GRMZM2G129907_T02;
Chr10 Ensembl_AGPv3 exon 1721994 1722055 . + .
Name=GRMZM2G129907_E05;Parent=GRMZM2G129907_T02;ensembl_end_phase=-1;ensembl_phase=-1;rank=2;ID=GRMZM2G129907_E05
Chr10 Ensembl_AGPv3 exon 1722296 1722779 . + .
Name=GRMZM2G129907_E01;Parent=GRMZM2G129907_T02;ensembl_end_phase=-1;ensembl_phase=-1;rank=3;ID=GRMZM2G129907_E01
Chr10 Ensembl_AGPv3 exon 1721887 1722055 . + .
Name=GRMZM2G129907_E07;Parent=GRMZM2G129907_T03;ensembl_end_phase=-1;ensembl_phase=-1;rank=1;ID=GRMZM2G129907_E07
Chr10 Ensembl_AGPv3 five_prime_UTR 1721887 1722363 . + .
Parent=GRMZM2G129907_T03;
Chr10 Ensembl_AGPv3 exon 1722296 1722779 . + .
Name=GRMZM2G129907_E01;Parent=GRMZM2G129907_T03;ensembl_end_phase=-1;ensembl_phase=-1;rank=2;ID=GRMZM2G129907_E01
Chr10 Ensembl_AGPv3 CDS 1722364 1722459 . + 0
Name=GRMZM2G129907_P03;Parent=GRMZM2G129907_T03;rank=1;ID=GRMZM2G129907_P03
Chr10 Ensembl_AGPv3 three_prime_UTR 1722460 1722779 . + .
Parent=GRMZM2G129907_T03;/
<Loading Image...
This is close to what I want, but the inner exons aren't showing. When I use
"sub_part=exon" in this track's stanza, the exons show but the gray UTRs
<Loading Image...
I want to render a display that looks exactly like the image above, but with
the grayed UTR regions showing in the exons. Any help on accomplishing this
would be greatly appreciated.
Thanks,
John
--
View this message in context: http://generic-model-organism-system-database.450254.n5.nabble.com/Configuring-Genes-to-show-exons-and-UTRs-tp5712467.html
Sent from the gmod-gbrowse mailing list archive at Nabble.com.